Symbiotic nitrogen fixation
The dependence of plants on metabolically usable nitrogen forms in soil limits their growth over time. High-yield agriculture depends on enriching soil with nitrogen, and this is achieved through the application of synthetic nitrogen fertilizers. Nevertheless, the excessive use of nitrogen fertilizers contributes to global warming and ozone depletion and leads to detrimental environmental pollution, such as contamination of ground water and the formation of hypoxic areas in oceans and lakes.
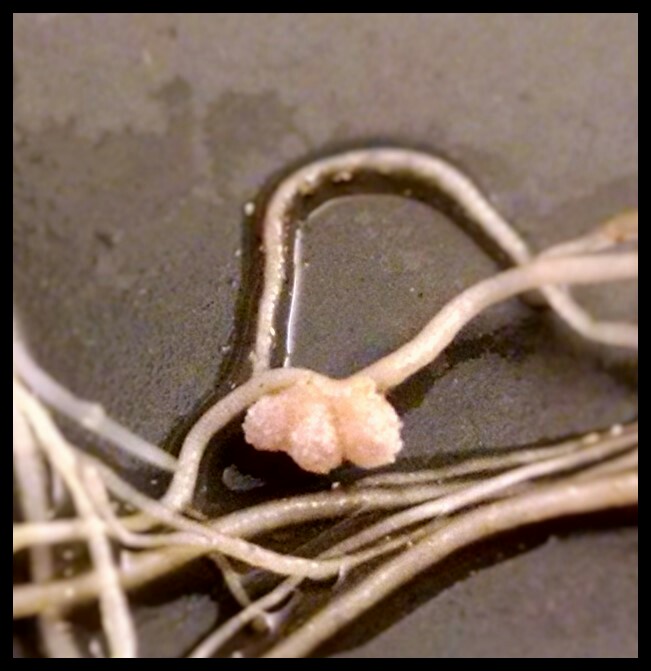
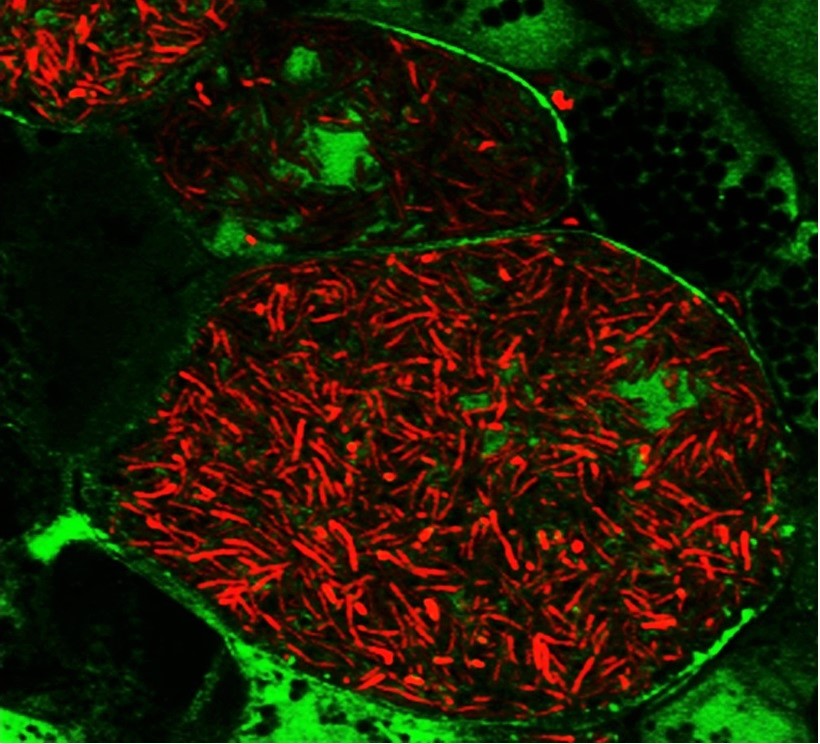
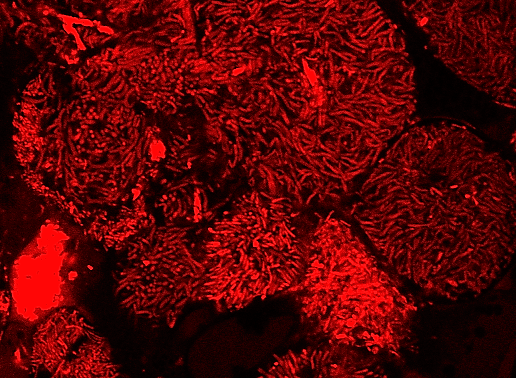
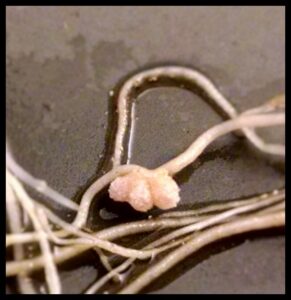
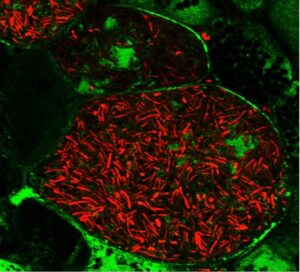
Legumes overcome nitrogen limitation by forming a symbiotic interaction with rhizobia, nitrogen-fixing soil bacteria that converts atmospheric nitrogen to usable nitrogen forms. This interaction occurs in a special organ in legumes’ roots, called nodule, in which bacteria produce ammonia and trade it with host plant for a carbon source and reducing power. In nodule cells, bacteria are internalized by host plant cells inside an intracellular compartment called the symbiosome and morphologically differentiate into nitrogen-fixing forms. One focus of our lab is illuminating the molecular mechanism of symbiotic nitrogen fixation by identifying the legume and rhizobia proteins involved in this process. By this way, we aim to improve the efficiency of nitrogen fixation in legumes and ultimately transfer the ability of forming symbiotic interaction with rhizobia to non-legume crops, thereby reduce or eliminate the need for synthetic nitrogen fertilizer in agriculture.
DNA damage and repair in plants
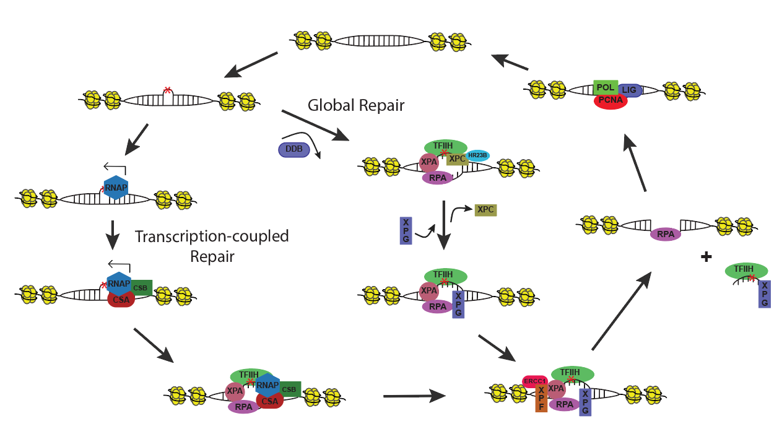
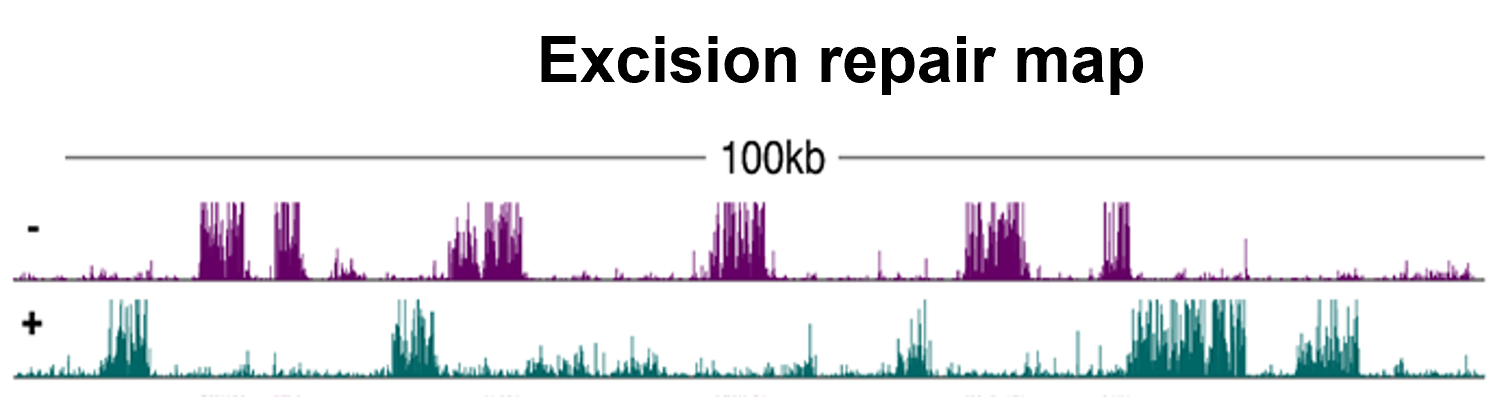
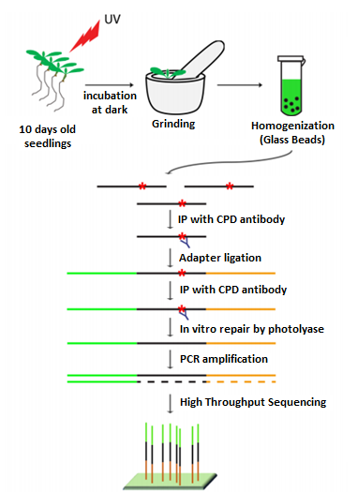
Plants are exposed to DNA-damaging environmental factors more than other organisms owing to their sessile lifestyles and dependence on sunlight for photosynthesis. However, the molecular mechanism of plant DNA repair mechanisms and their genome-wide dynamics and influencers are relatively less understood. In our lab, we are working on these concepts and developing approaches to improve genotoxic stress tolerance in plants. We are focusing on plant nucleotide excision repair that eliminates transcription- and replication-blocking bulky DNA adducts. NER detects bulky DNA adducts and removes the adduct-containing oligomers by properly concerted dual (5’ and 3’) incision, followed by gap filling and ligation. NER is stimulated either by the direct recognition of DNA adducts, called global repair, or by the DNA damage-blocked RNA polymerase II, named transcription-coupled repair (TCR). While TCR occurs only in transcribed regions, global repair proceeds in non-transcribed and intergenic regions of the genome. Using the XR-seq method, we generated the genome-wide repair map of UV damage at single nucleotide resolution in Arabidopsis thaliana at different timepoints of a day. We found that the chromatin state influences repair rate, however the main determinant of repair rate is transcription. In our group, we are working on understanding the molecular mechanism of NER in plants, identifying novel DNA-damaging agents with a focus on microbial substances and explaining genome-wide dynamics of NER.